homogenization/perfusion_micro.py¶
Description
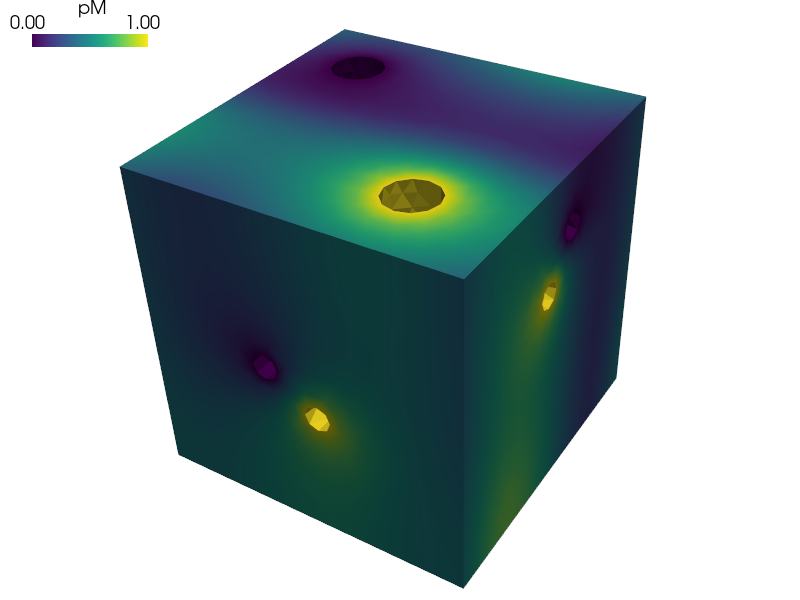
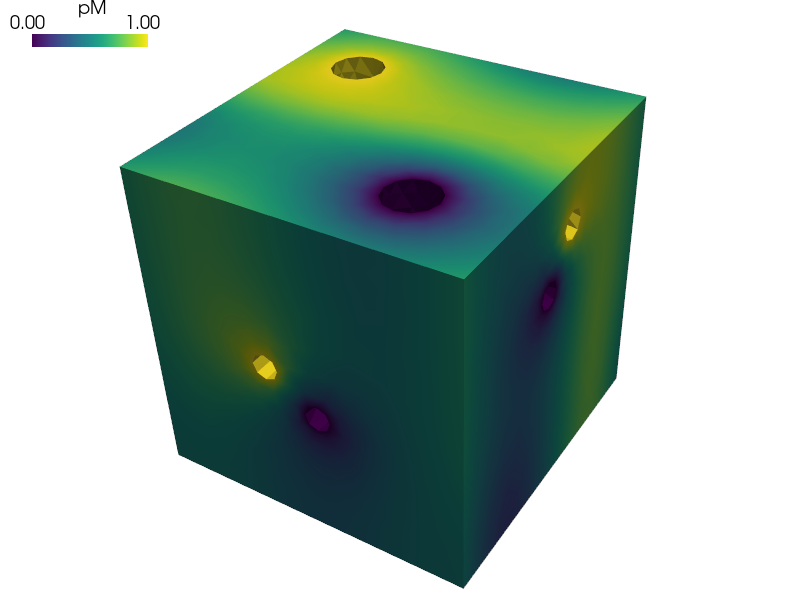
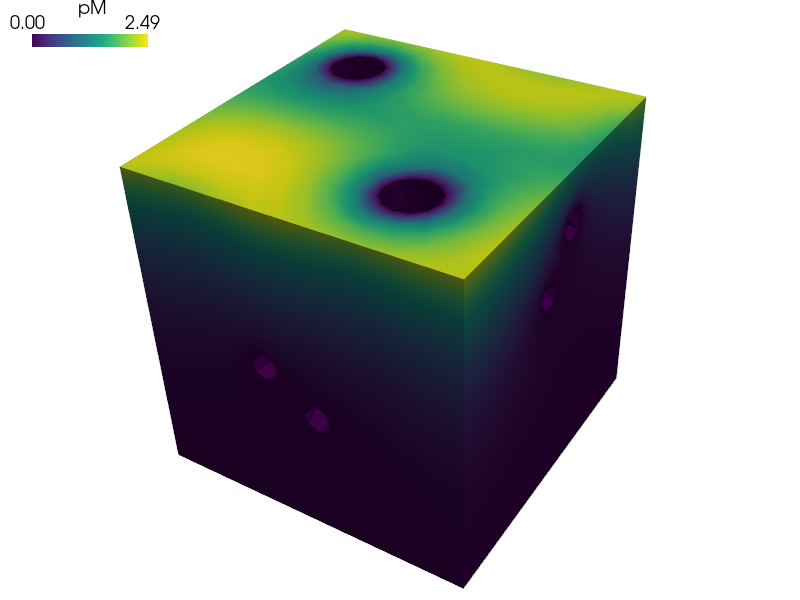
Homogenization of the Darcy flow in a thin porous layer.
The reference cell is composed of the matrix representing the dual porosity and of two disconnected channels representing the primary porosity, see paper [1].
[1] E. Rohan, V. Lukeš: Modeling Tissue Perfusion Using a Homogenized Model with Layer-wise Decomposition. IFAC Proceedings Volumes 45(2), 2012, pages 1029-1034. https://doi.org/10.3182/20120215-3-AT-3016.00182
# -*- coding: utf-8
r"""
Homogenization of the Darcy flow in a thin porous layer.
The reference cell is composed of the matrix representing the dual porosity
and of two disconnected channels representing the primary porosity,
see paper [1].
[1] E. Rohan, V. Lukeš: Modeling Tissue Perfusion Using a Homogenized
Model with Layer-wise Decomposition. IFAC Proceedings Volumes 45(2), 2012,
pages 1029-1034.
https://doi.org/10.3182/20120215-3-AT-3016.00182
"""
from __future__ import absolute_import
from sfepy.discrete.fem.periodic import match_x_plane, match_y_plane
import sfepy.homogenization.coefs_base as cb
import numpy as nm
from sfepy import data_dir
import six
from six.moves import range
def get_mats(pk, ph, pe, dim):
m1 = nm.eye(dim, dtype=nm.float64) * pk
m1[-1, -1] = pk / ph
m2 = nm.eye(dim, dtype=nm.float64) * pk
m2[-1, -1] = pk / ph ** 2
return m1, m2
def recovery_perf(pb, corrs, macro):
from sfepy.homogenization.recovery import compute_p_from_macro
from sfepy.base.base import Struct
slev = ''
micro_nnod = pb.domain.mesh.n_nod
centre_Y = nm.sum(pb.domain.mesh.coors, axis=0) / micro_nnod
nodes_Y = {}
channels = {}
for k in six.iterkeys(macro):
if 'press' in k:
channels[k[-1]] = 1
channels = list(channels.keys())
varnames = ['pM']
for ch in channels:
nodes_Y[ch] = pb.domain.regions['Y' + ch].vertices
varnames.append('p' + ch)
pvars = pb.create_variables(varnames)
press = {}
# matrix
press['M'] = \
corrs['corrs_%s_gamma_p' % pb_def['name']]['pM'] * macro['g_p'] + \
corrs['corrs_%s_gamma_m' % pb_def['name']]['pM'] * macro['g_m']
out = {}
# channels
for ch in channels:
press_mac = macro['press' + ch][0, 0]
press_mac_grad = macro['pressg' + ch]
nnod = corrs['corrs_%s_pi%s' % (pb_def['name'], ch)]\
['p%s_0' % ch].shape[0]
press_mic = nm.zeros((nnod, 1))
for key, val in \
six.iteritems(corrs['corrs_%s_pi%s' % (pb_def['name'], ch)]):
kk = int(key[-1])
press_mic += val * press_mac_grad[kk, 0]
for key in six.iterkeys(corrs):
if ('_gamma_' + ch in key):
kk = int(key[-1]) - 1
press_mic += corrs[key]['p' + ch] * macro['g' + ch][kk]
press_mic += \
compute_p_from_macro(press_mac_grad[nm.newaxis,nm.newaxis, :, :],
micro_coors[nodes_Y[ch]], 0,
centre=centre_Y, extdim=-1).reshape((nnod, 1))
press[ch] = press_mac + eps0 * press_mic
out[slev + 'p' + ch] = Struct(name='output_data',
mode='vertex',
data=press[ch],
var_name='p' + ch,
dofs=None)
pvars['p' + ch].set_data(press_mic)
dvel = pb.evaluate('ev_diffusion_velocity.iV.Y%s(mat1%s.k, p%s)'
% (ch, ch, ch),
var_dict={'p' + ch: pvars['p' + ch]},
mode='el_avg')
out[slev + 'w' + ch] = Struct(name='output_data',
mode='cell',
data=dvel,
var_name='w' + ch,
dofs=None)
press['M'] += corrs['corrs_%s_eta%s' % (pb_def['name'], ch)]['pM']\
* press_mac
pvars['pM'].set_data(press['M'])
dvel = pb.evaluate('%e * ev_diffusion_velocity.iV.YM(mat1M.k, pM)' % eps0,
var_dict={'pM': pvars['pM']}, mode='el_avg')
out[slev + 'pM'] = Struct(name='output_data',
mode='vertex',
dat=press['M'],
var_name='pM',
dofs=None)
out[slev + 'wM'] = Struct(name='output_data',
mode='cell',
data=dvel,
var_name='wM',
dofs=None)
return out
geoms = {
'2_4': ['2_4_Q1', '2', 5],
'3_8': ['3_8_Q1', '4', 5],
'3_4': ['3_4_P1', '3', 3],
}
pb_def = {
'name': '3d_2ch',
'mesh_filename': data_dir + '/meshes/3d/perfusion_micro3d.mesh',
'dim': 3,
'geom': geoms['3_4'],
'eps0': 1.0e-2,
'param_h': 1.0,
'param_kappa_m': 0.1,
'matrix_mat_el_grp': 3,
'channels': {
'A': {
'mat_el_grp': 1,
'fix_nd_grp': (4, 1),
'io_nd_grp': [1, 2, 3],
'param_kappa_ch': 1.0,
},
'B': {
'mat_el_grp': 2,
'fix_nd_grp': (14, 11),
'io_nd_grp': [11, 12, 13],
'param_kappa_ch': 2.0,
},
},
}
filename_mesh = pb_def['mesh_filename']
eps0 = pb_def['eps0']
param_h = pb_def['param_h']
# integrals
integrals = {
'iV': 2,
'iS': 2,
}
functions = {
'match_x_plane': (match_x_plane,),
'match_y_plane': (match_y_plane,),
}
aux = []
for ch, val in six.iteritems(pb_def['channels']):
aux.append('r.bYM' + ch)
# basic regions
regions = {
'Y': 'all',
'YM': 'cells of group %d' % pb_def['matrix_mat_el_grp'],
# periodic boundaries
'Pl': ('vertices in (x < 0.001)', 'facet'),
'Pr': ('vertices in (x > 0.999)', 'facet'),
'PlYM': ('r.Pl *v r.YM', 'facet'),
'PrYM': ('r.Pr *v r.YM', 'facet'),
'bYMp': ('r.bYp *v r.YM', 'facet', 'YM'),
'bYMm': ('r.bYm *v r.YM', 'facet', 'YM'),
'bYMpm': ('r.bYMp +v r.bYMm', 'facet', 'YM'),
}
# matrix/channel boundaries
regions.update({
'bYMchs': (' +v '.join(aux), 'facet', 'YM'),
'YMmchs': 'r.YM -v r.bYMchs',
})
# boundary conditions Gamma+/-
ebcs = {
'gamma_pm_bYMchs': ('bYMchs', {'pM.0': 0.0}),
'gamma_pm_YMmchs': ('YMmchs', {'pM.0': 1.0}),
}
# periodic boundary conditions - matrix, X-direction
epbcs = {'periodic_xYM': (['PlYM', 'PrYM'], {'pM.0': 'pM.0'}, 'match_x_plane')}
lcbcs = {}
all_periodicYM = ['periodic_%sYM' % ii for ii in ['x', 'y'][:pb_def['dim']-1]]
all_periodicY = {}
if pb_def['dim'] == 2:
regions.update({
'bYm': ('vertices in (y < 0.001)', 'facet'),
'bYp': ('vertices in (y > 0.999)', 'facet'),
})
if pb_def['dim'] == 3:
regions.update({
'Pn': ('vertices in (y < 0.001)', 'facet'),
'Pf': ('vertices in (y > 0.999)', 'facet'),
'PnYM': ('r.Pn *v r.YM', 'facet'),
'PfYM': ('r.Pf *v r.YM', 'facet'),
'bYm': ('vertices in (z < 0.001)', 'facet'),
'bYp': ('vertices in (z > 0.999)', 'facet'),
})
# periodic boundary conditions - matrix, Y-direction
epbcs.update({
'periodic_yYM': (['PnYM', 'PfYM'], {'pM.0': 'pM.0'}, 'match_y_plane'),
})
reg_io = {}
ebcs_eta = {}
ebcs_gamma = {}
# generate regions, ebcs, epbcs
for ch, val in six.iteritems(pb_def['channels']):
all_periodicY[ch] = ['periodic_%sY%s' % (ii, ch)
for ii in ['x', 'y'][:pb_def['dim']-1]]
# channels: YA, fixedYA, bYMA (matrix/channel boundaries)
regions.update({
'Y' + ch: 'cells of group %d' % val['mat_el_grp'],
'bYM' + ch: ('r.YM *v r.Y' + ch, 'facet', 'YM'),
'PlY' + ch: ('r.Pl *v r.Y' + ch, 'facet'),
'PrY' + ch: ('r.Pr *v r.Y' + ch, 'facet'),
})
if 'fix_nd_grp' in val:
regions.update({
'fixedY' + ch: ('vertices of group %d' % val['fix_nd_grp'][0],
'vertex'),
})
ebcs_eta[ch] = []
for ch2, val2 in six.iteritems(pb_def['channels']):
aux = 'eta%s_bYM%s' % (ch, ch2)
if ch2 == ch:
ebcs.update({aux: ('bYM' + ch2, {'pM.0': 1.0})})
else:
ebcs.update({aux: ('bYM' + ch2, {'pM.0': 0.0})})
ebcs_eta[ch].append(aux)
# boundary conditions
# periodic boundary conditions - channels, X-direction
epbcs.update({
'periodic_xY' + ch: (['PlY' + ch, 'PrY' + ch],
{'p%s.0' % ch: 'p%s.0' % ch},
'match_x_plane'),
})
if pb_def['dim'] == 3:
regions.update({
'PnY' + ch: ('r.Pn *v r.Y' + ch, 'facet'),
'PfY' + ch: ('r.Pf *v r.Y' + ch, 'facet'),
})
# periodic boundary conditions - channels, Y-direction
epbcs.update({
'periodic_yY' + ch: (['PnY' + ch, 'PfY' + ch],
{'p%s.0' % ch: 'p%s.0' % ch},
'match_y_plane'),
})
reg_io[ch] = []
aux_bY = []
# channel: inputs/outputs
for i_io in range(len(val['io_nd_grp'])):
io = '%s_%d' % (ch, i_io+1)
# regions
aux = val['io_nd_grp'][i_io]
if 'fix_nd_grp' in val and val['fix_nd_grp'][1] == aux:
regions.update({
'bY%s' % io: ('vertices of group %d +v r.fixedY%s' % (aux, ch),
'facet', 'Y%s' % ch),
})
else:
regions.update({
'bY%s' % io: ('vertices of group %d' % aux,
'facet', 'Y%s' % ch),
})
aux_bY.append('r.bY%s' % io)
reg_io[ch].append('bY%s' % io)
regions.update({
'bY' + ch: (' +v '.join(aux_bY), 'facet', 'Y' + ch),
})
# channel: inputs/outputs
for i_io in range(len(val['io_nd_grp'])):
io = '%s_%d' % (ch, i_io + 1)
ion = '%s_n%d' % (ch, i_io + 1)
regions.update({
'bY%s' % ion: ('r.bY%s -v r.bY%s' % (ch, io), 'facet', 'Y%s' % ch),
})
# boundary conditions
aux = 'fix_p%s_bY%s' % (ch, ion)
ebcs.update({
aux: ('bY%s' % ion, {'p%s.0' % ch: 0.0}),
})
lcbcs.update({
'imv' + ch: ('Y' + ch, {'ls%s.all' % ch: None}, None,
'integral_mean_value')
})
matk1, matk2 = get_mats(pb_def['param_kappa_m'], param_h, eps0, pb_def['dim'])
materials = {
'mat1M': ({'k': matk1},),
'mat2M': ({'k': matk2},),
}
fields = {
'corrector_M': ('real', 'scalar', 'YM', 1),
'vel_M': ('real', 'vector', 'YM', 1),
'vol_all': ('real', 'scalar', 'Y', 1),
}
variables = {
'pM': ('unknown field', 'corrector_M'),
'qM': ('test field', 'corrector_M', 'pM'),
'Pi_M': ('parameter field', 'corrector_M', '(set-to-None)'),
'corr_M': ('parameter field', 'corrector_M', '(set-to-None)'),
'corr1_M': ('parameter field', 'corrector_M', '(set-to-None)'),
'corr2_M': ('parameter field', 'corrector_M', '(set-to-None)'),
'wM': ('parameter field', 'vel_M', '(set-to-None)'),
'vol_all': ('parameter field', 'vol_all', '(set-to-None)'),
}
# generate regions for channel inputs/outputs
for ch, val in six.iteritems(pb_def['channels']):
matk1, matk2 = get_mats(val['param_kappa_ch'], param_h,
eps0, pb_def['dim'])
materials.update({
'mat1' + ch: ({'k': matk1},),
'mat2' + ch: ({'k': matk2},),
})
fields.update({
'corrector_' + ch: ('real', 'scalar', 'Y' + ch, 1),
'vel_' + ch: ('real', 'vector', 'Y' + ch, 1),
})
variables.update({
'p' + ch: ('unknown field', 'corrector_' + ch),
'q' + ch: ('test field', 'corrector_' + ch, 'p' + ch),
'Pi_' + ch: ('parameter field', 'corrector_' + ch, '(set-to-None)'),
'corr1_' + ch: ('parameter field', 'corrector_' + ch, '(set-to-None)'),
'corr2_' + ch: ('parameter field', 'corrector_' + ch, '(set-to-None)'),
'w' + ch: ('unknown field', 'vel_' + ch),
# lagrange mutltipliers - integral mean value
'ls' + ch: ('unknown field', 'corrector_' + ch),
'lv' + ch: ('test field', 'corrector_' + ch, 'ls' + ch),
})
options = {
'coefs': 'coefs',
'requirements': 'requirements',
'ls': 'ls', # linear solver to use
'volumes': {
'total': {
'variables': ['vol_all'],
'expression': """ev_volume.iV.Y(vol_all)""",
},
'one': {
'value': 1.0,
}
},
'output_dir': './output',
'split_results_by': 'region',
'coefs_filename': 'coefs_perf_' + pb_def['name'],
'coefs_info': {'eps0': eps0},
'recovery_hook': 'recovery_perf',
'multiprocessing': False,
}
for ipm in ['p', 'm']:
options['volumes'].update({
'bYM' + ipm: {
'variables': ['pM'],
'expression': "ev_volume.iS.bYM%s(pM)" % ipm,
},
'bY' + ipm: {
'variables': ['vol_all'],
'expression': "ev_volume.iS.bY%s(vol_all)" % ipm,
}
})
for ch in six.iterkeys(reg_io):
for ireg in reg_io[ch]:
options['volumes'].update({
ireg: {
'variables': ['p' + ch],
'expression': "ev_volume.iS.%s(p%s)" % (ireg, ch),
}
})
coefs = {
'vol_bYMpm': {
'regions': ['bYMp', 'bYMm'],
'expression': 'ev_volume.iS.%s(pM)',
'class': cb.VolumeFractions,
},
'filenames': {},
}
requirements = {
'corrs_one_YM': {
'variable': ['pM'],
'ebcs': ['gamma_pm_YMmchs', 'gamma_pm_bYMchs'],
'epbcs': [],
'save_name': 'corrs_one_YM',
'class': cb.CorrSetBCS,
'is_linear': True,
},
}
for ipm in ['p', 'm']:
requirements.update({
'corrs_gamma_' + ipm: {
'requires': [],
'ebcs': ['gamma_pm_bYMchs'],
'epbcs': all_periodicYM,
'equations': {
'eq_gamma_pm': """dw_diffusion.iV.YM(mat2M.k, qM, pM) =
%e * dw_integrate.iS.bYM%s(qM)"""
% (1.0/param_h, ipm),
},
'class': cb.CorrOne,
'save_name': 'corrs_%s_gamma_%s' % (pb_def['name'], ipm),
'is_linear': True,
},
})
for ipm2 in ['p', 'm']:
coefs.update({
'H' + ipm + ipm2: { # test+
'requires': ['corrs_gamma_' + ipm],
'set_variables': [('corr_M', 'corrs_gamma_' + ipm, 'pM')],
'expression': 'ev_integrate.iS.bYM%s(corr_M)' % ipm2,
'set_volume': 'bYp',
'class': cb.CoefOne,
},
})
def get_channel(keys, bn):
for ii in keys:
if bn in ii:
return ii[(ii.rfind(bn) + len(bn)):]
return None
def set_corrpis(variables, ir, ic, mode, **kwargs):
ch = get_channel(list(kwargs.keys()), 'pis_')
pis = kwargs['pis_' + ch]
corrs_pi = kwargs['corrs_pi' + ch]
if mode == 'row':
val = pis.states[ir]['p' + ch] + corrs_pi.states[ir]['p' + ch]
variables['corr1_' + ch].set_data(val)
elif mode == 'col':
val = pis.states[ic]['p' + ch] + corrs_pi.states[ic]['p' + ch]
variables['corr2_' + ch].set_data(val)
def set_corr_S(variables, ir, *args, **kwargs):
ch = get_channel(list(kwargs.keys()), 'pis_')
io = get_channel(list(kwargs.keys()), 'corrs_gamma_')
pis = kwargs['pis_' + ch]
corrs_gamma = kwargs['corrs_gamma_' + io]
pi = pis.states[ir]['p' + ch]
val = corrs_gamma.state['p' + ch]
variables['corr1_' + ch].set_data(pi)
variables['corr2_' + ch].set_data(val)
def set_corr_cc(variables, ir, *args, **kwargs):
ch = get_channel(list(kwargs.keys()), 'pis_')
pis = kwargs['pis_' + ch]
corrs_pi = kwargs['corrs_pi' + ch]
pi = pis.states[ir]['p' + ch]
pi = pi - nm.mean(pi)
val = pi + corrs_pi.states[ir]['p' + ch]
variables['corr1_' + ch].set_data(val)
for ch, val in six.iteritems(pb_def['channels']):
coefs.update({
'G' + ch: { # test+
'requires': ['corrs_one' + ch, 'corrs_eta' + ch],
'set_variables': [('corr1_M', 'corrs_one' + ch, 'pM'),
('corr2_M', 'corrs_eta' + ch, 'pM')],
'expression': 'dw_diffusion.iV.YM(mat2M.k, corr1_M, corr2_M)',
'class': cb.CoefOne,
},
'K' + ch: { # test+
'requires': ['pis_' + ch, 'corrs_pi' + ch],
'set_variables': set_corrpis,
'expression': 'dw_diffusion.iV.Y%s(mat2%s.k, corr1_%s, corr2_%s)'\
% ((ch,) * 4),
'dim': pb_def['dim'] - 1,
'class': cb.CoefDimDim,
},
})
requirements.update({
'pis_' + ch: {
'variables': ['p' + ch],
'class': cb.ShapeDim,
},
'corrs_one' + ch: {
'variable': ['pM'],
'ebcs': ebcs_eta[ch],
'epbcs': [],
'save_name': 'corrs_%s_one%s' % (pb_def['name'], ch),
'class': cb.CorrSetBCS,
},
'corrs_eta' + ch: {
'ebcs': ebcs_eta[ch],
'epbcs': all_periodicYM,
'equations': {
'eq_eta': 'dw_diffusion.iV.YM(mat2M.k, qM, pM) = 0',
},
'class': cb.CorrOne,
'save_name': 'corrs_%s_eta%s' % (pb_def['name'], ch),
'is_linear': True,
},
'corrs_pi' + ch: {
'requires': ['pis_' + ch],
'set_variables': [('Pi_' + ch, 'pis_' + ch, 'p' + ch)],
'ebcs': [],
'epbcs': all_periodicY[ch],
'lcbcs': ['imv' + ch],
'equations': {
'eq_pi': """dw_diffusion.iV.Y%s(mat2%s.k, q%s, p%s)
+ dw_dot.iV.Y%s(q%s, ls%s)
= - dw_diffusion.iV.Y%s(mat2%s.k, q%s, Pi_%s)"""
% ((ch,) * 11),
'eq_imv': 'dw_dot.iV.Y%s(lv%s, p%s) = 0' % ((ch,) * 3),
},
'dim': pb_def['dim'] - 1,
'class': cb.CorrDim,
'save_name': 'corrs_%s_pi%s' % (pb_def['name'], ch),
},
})
for ipm in ['p', 'm']:
coefs.update({
'E' + ipm + ch: { # test+
'requires': ['corrs_eta' + ch],
'set_variables': [('corr_M', 'corrs_eta' + ch, 'pM')],
'expression': 'ev_integrate.iS.bYM%s(corr_M)' % ipm,
'set_volume': 'bYp',
'class': cb.CoefOne,
},
'F' + ipm + ch: { # test+
'requires': ['corrs_one' + ch, 'corrs_gamma_' + ipm],
'set_variables': [('corr1_M', 'corrs_one' + ch, 'pM'),
('corr2_M', 'corrs_gamma_' + ipm, 'pM')],
'expression': """dw_diffusion.iV.YM(mat2M.k, corr1_M, corr2_M)
- %e * ev_integrate.iS.bYM%s(corr1_M)"""\
% (1.0/param_h, ipm),
'class': cb.CoefOne,
},
})
for i_io in range(len(val['io_nd_grp'])):
io = '%s_%d' % (ch, i_io + 1)
coefs.update({
'S' + io: { # [Rohan1] (4.28), test+
'requires': ['corrs_gamma_' + io, 'pis_' + ch],
'set_variables': set_corr_S,
'expression': 'dw_diffusion.iV.Y%s(mat2%s.k,corr1_%s,corr2_%s)'
% ((ch,) * 4),
'dim': pb_def['dim'] - 1,
'class': cb.CoefDim,
},
'P' + io: { # test+
'requires': ['pis_' + ch, 'corrs_pi' + ch],
'set_variables': set_corr_cc,
'expression': 'ev_integrate.iS.bY%s(corr1_%s)'\
% (io, ch),
'set_volume': 'bYp',
'dim': pb_def['dim'] - 1,
'class': cb.CoefDim,
},
'S_test' + io: {
'requires': ['corrs_pi' + ch],
'set_variables': [('corr1_' + ch, 'corrs_pi' + ch, 'p' + ch)],
'expression': '%e * ev_integrate.iS.bY%s(corr1_%s)'\
% (1.0 / param_h, io, ch),
'dim': pb_def['dim'] - 1,
'class': cb.CoefDim,
},
})
requirements.update({
'corrs_gamma_' + io: {
'requires': [],
'variables': ['p' + ch, 'q' + ch],
'ebcs': [],
'epbcs': all_periodicY[ch],
'lcbcs': ['imv' + ch],
'equations': {
'eq_gamma': """dw_diffusion.iV.Y%s(mat2%s.k, q%s, p%s)
+ dw_dot.iV.Y%s(q%s, ls%s)
= %e * dw_integrate.iS.bY%s(q%s)"""
% ((ch,) * 7 + (1.0/param_h, io, ch)),
'eq_imv': 'dw_dot.iV.Y%s(lv%s, p%s) = 0'
% ((ch,) * 3),
},
'class': cb.CorrOne,
'save_name': 'corrs_%s_gamma_%s' % (pb_def['name'], io),
},
})
for i_io2 in range(len(val['io_nd_grp'])):
io2 = '%s_%d' % (ch, i_io2 + 1)
io12 = '%s_%d' % (io, i_io2 + 1)
coefs.update({
'R' + io12: { # test+
'requires': ['corrs_gamma_' + io2],
'set_variables': [('corr1_' + ch, 'corrs_gamma_' + io2,
'p' + ch)],
'expression': 'ev_integrate.iS.bY%s(corr1_%s)'\
% (io, ch),
'set_volume': 'bYp',
'class': cb.CoefOne,
},
})
solvers = {
'ls': ('ls.auto_direct', {'use_presolve' : True}),
'newton': ('nls.newton', {
'i_max': 1,
})
}