homogenization/linear_elastic_mM.py¶
Description
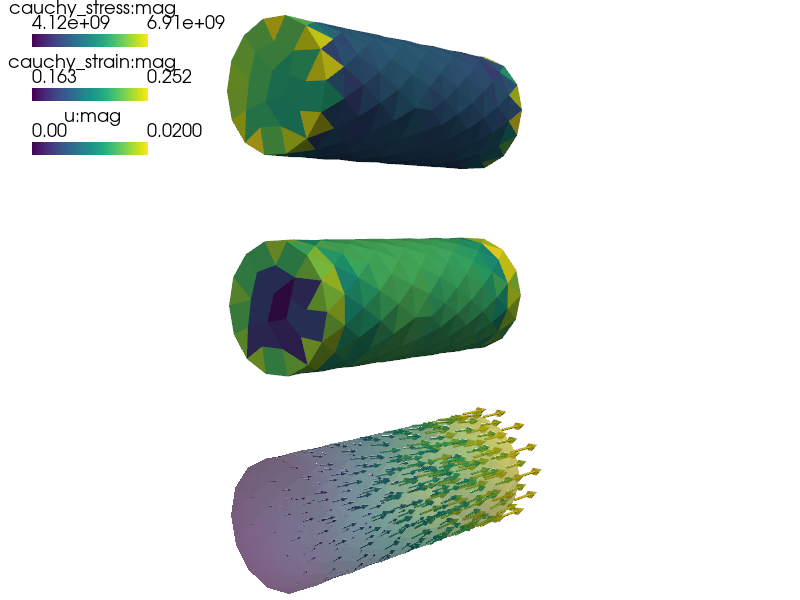
Linear elasticity with effective macroscopic properties determined according to the theory of homogenization from a periodic microstructure.
"""
Linear elasticity with effective macroscopic properties determined
according to the theory of homogenization from a periodic microstructure.
"""
import os
from sfepy import data_dir, base_dir
from sfepy.base.base import nm
from sfepy.homogenization.micmac import get_homog_coefs_linear
from sfepy.homogenization.recovery import save_recovery_region,\
recover_micro_hook
def post_process(out, pb, state, extend=False):
from sfepy.base.base import Struct
if isinstance(state, dict):
pass
else:
stress = pb.evaluate('ev_cauchy_stress.i.Omega(solid.D, u)',
mode='el_avg')
strain = pb.evaluate('ev_cauchy_strain.i.Omega(u)',
mode='el_avg')
out['cauchy_strain'] = Struct(name='output_data',
mode='cell', data=strain,
dofs=None)
out['cauchy_stress'] = Struct(name='output_data',
mode='cell', data=stress,
dofs=None)
if pb.conf.options.get('recover_micro', False):
rname = pb.conf.options.recovery_region
region = pb.domain.regions[rname]
filename = os.path.join(os.path.dirname(pb.get_output_name()),
'recovery_region.vtk')
save_recovery_region(pb, rname, filename=filename);
rstrain = pb.evaluate('ev_cauchy_strain.i.%s(u)' % rname,
mode='el_avg')[:, 0, ...]
recover_micro_hook(pb.conf.options.micro_filename,
region, {'strain': rstrain}, 0.01,
output_dir=pb.conf.options.output_dir)
return out
def get_elements(coors, domain=None):
return nm.arange(50, domain.shape.n_el, 100)
regenerate = True
def get_homog(ts, coors, mode=None,
equations=None, term=None, problem=None, **kwargs):
global regenerate
out = get_homog_coefs_linear(ts, coors, mode, regenerate=regenerate,
micro_filename=options['micro_filename'],
output_dir=problem.conf.options.output_dir)
regenerate = False
return out
functions = {
'get_elements' : (get_elements,),
'get_homog' : (get_homog,),
}
filename_mesh = data_dir + '/meshes/3d/cylinder.mesh'
regions = {
'Omega' : 'all',
'Left' : ('vertices in (x < 0.001)', 'facet'),
'Right' : ('vertices in (x > 0.099)', 'facet'),
'Recovery' : 'cells by get_elements',
}
materials = {
'solid' : 'get_homog',
}
fields = {
'3_displacement' : ('real', 3, 'Omega', 1),
}
integrals = {
'i' : 1,
}
variables = {
'u' : ('unknown field', '3_displacement', 0),
'v' : ('test field', '3_displacement', 'u'),
}
ebcs = {
'Fixed' : ('Left', {'u.all' : 0.0}),
'PerturbedSurface' : ('Right', {'u.0' : 0.02, 'u.1' : 0.0, 'u.2' : 0.0}),
}
equations = {
'balance_of_forces' :
"""dw_lin_elastic.i.Omega(solid.D, v, u) = 0""",
}
solvers = {
'ls' : ('ls.scipy_direct', {}),
'newton' : ('nls.newton', {
'i_max' : 1,
'eps_a' : 1e-6,
}),
}
micro_filename = base_dir \
+ '/examples/homogenization/linear_homogenization_up.py'
options = {
'nls' : 'newton',
'ls' : 'ls',
'output_dir' : 'output',
'post_process_hook' : 'post_process',
'output_prefix' : 'macro:',
'recover_micro': True,
'recovery_region' : 'Recovery',
'micro_filename' : micro_filename,
}