dg/burgers_2D.py¶
Description
Burgers equation in 2D solved using discontinous Galerkin method
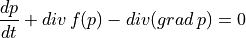
Based on
Kučera, V. (n.d.). Higher order methods for the solution of compressible flows. Charles University. p. 21 eq. (1.39)
Usage Examples¶
Run:
sfepy-run sfepy/examples/dg/burgers_2D.py
Results are saved to output/dg/burgers_2D folder by default as .msh files,
the best way to view them is through GMSH (http://gmsh.info/) version 4.6 or
newer. Start GMSH and use File | Open menu or Crtl + O shortcut, navigate to
the output folder, select all .msh files and hit Open, all files should load
as one item in Post-processing named p_cell_nodes.
GMSH is capable of rendering high order approximations in individual elements,
to modify fidelity of rendering, double click the displayed mesh, quick options
menu should pop up, click on All view options.... This brings up the Options
window with View [0] selected in left column. Under the tab General
ensure that Adapt visualization grid is ticked, then you can adjust
Maximum recursion depth and `Target visualization error to tune
the visualization. To see visualization elements (as opposed to mesh elements)
go to Visibility tab and tick Draw element outlines, this option is also
available from quick options menu as View element outlines or under shortcut
Alt+E. In the quick options menu, you can also modify normal raise by
clicking View Normal Raise to see solution rendered as surface above the
mesh. Note that for triangular meshes normal raise -1 produces expected raise
above the mesh. This is due to the opposite orientation of the reference
elements in GMSH and Sfepy and might get patched in the future.
r"""
Burgers equation in 2D solved using discontinous Galerkin method
.. math:: \frac{dp}{dt} + div\,f(p) - div(grad\,p) = 0
Based on
Kučera, V. (n.d.). Higher order methods for the solution of compressible flows.
Charles University. p. 21 eq. (1.39)
Usage Examples
--------------
Run::
sfepy-run sfepy/examples/dg/burgers_2D.py
Results are saved to output/dg/burgers_2D folder by default as ``.msh`` files,
the best way to view them is through GMSH (http://gmsh.info/) version 4.6 or
newer. Start GMSH and use ``File | Open`` menu or Crtl + O shortcut, navigate to
the output folder, select all ``.msh`` files and hit Open, all files should load
as one item in Post-processing named p_cell_nodes.
GMSH is capable of rendering high order approximations in individual elements,
to modify fidelity of rendering, double click the displayed mesh, quick options
menu should pop up, click on ``All view options...``. This brings up the Options
window with ``View [0]`` selected in left column. Under the tab ``General``
ensure that ``Adapt visualization grid`` is ticked, then you can adjust
``Maximum recursion depth`` and ```Target visualization error`` to tune
the visualization. To see visualization elements (as opposed to mesh elements)
go to ``Visibility`` tab and tick ``Draw element outlines``, this option is also
available from quick options menu as ``View element outlines`` or under shortcut
``Alt+E``. In the quick options menu, you can also modify normal raise by
clicking ``View Normal Raise`` to see solution rendered as surface above the
mesh. Note that for triangular meshes normal raise -1 produces expected raise
above the mesh. This is due to the opposite orientation of the reference
elements in GMSH and Sfepy and might get patched in the future.
"""
from sfepy.examples.dg.example_dg_common import *
from sfepy import data_dir
from sfepy.discrete.dg.limiters import MomentLimiter2D, IdentityLimiter
mesh_center = (0, 0)
mesh_size = (2, 2)
def define(filename_mesh=None,
approx_order=2,
adflux=0,
limit=False,
cw=10,
diffcoef=0.002,
diffscheme="symmetric",
cfl=None,
dt=1e-5,
t1=0.01
):
functions = {}
def local_register_function(fun):
try:
functions.update({fun.__name__: (fun,)})
except AttributeError: # Already a sfepy Function.
fun = fun.function
functions.update({fun.__name__: (fun,)})
return fun
example_name = "burgers_2D"
dim = 2
if filename_mesh is None:
filename_mesh = data_dir + "/meshes/2d/square_tri2.mesh"
t0 = 0.
if dt is None and cfl is None:
dt = 1e-5
velo = [1., 1.]
angle = 0 # - nm.pi / 5
rotm = nm.array([[nm.cos(angle), -nm.sin(angle)],
[nm.sin(angle), nm.cos(angle)]])
velo = nm.sum(rotm.T * nm.array(velo), axis=-1)[:, None]
burg_velo = velo.T / nm.linalg.norm(velo)
regions = {
'Omega': 'all',
'left' : ('vertices in x == -1', 'edge'),
'right': ('vertices in x == 1', 'edge'),
'top' : ('vertices in y == 1', 'edge'),
'bottom': ('vertices in y == -1', 'edge')
}
fields = {
'f': ('real', 'scalar', 'Omega',
str(approx_order) + 'd', 'DG', 'legendre')
}
variables = {
'p': ('unknown field', 'f', 0, 1),
'v': ('test field', 'f', 'p'),
}
integrals = {
'i': 5,
}
def analytic_sol(coors, t):
x_1 = coors[..., 0]
x_2 = coors[..., 1]
sin = nm.sin
pi = nm.pi
exp = nm.exp
res = -(exp(-t) - 1)*(sin(5*x_1*x_2) + sin(-4*x_1*x_2 + 4*x_1 + 4*x_2))
return res
@local_register_function
def sol_fun(ts, coors, mode="qp", **kwargs):
t = ts.time
if mode == "qp":
return {"p": analytic_sol(coors, t)[..., None, None]}
@local_register_function
def bc_funs(ts, coors, bc, problem):
# return 2*coors[..., 1]
t = ts.dt*ts.step
x_1 = coors[..., 0]
x_2 = coors[..., 1]
sin = nm.sin
cos = nm.cos
exp = nm.exp
if bc.diff == 0:
if "left" in bc.name:
res = -(exp(-t) - 1)*(sin(-5*x_2) + sin(8*x_2 - 4))
elif "bottom" in bc.name:
res = -(exp(-t) - 1) * (sin(-5 * x_1) + sin(8 * x_1 - 4))
elif "right" in bc.name:
res = -(exp(-t) - 1)*(sin(4) + sin(5*x_2))
elif "top" in bc.name:
res = -(exp(-t) - 1)*(sin(4) + sin(5*x_1))
elif bc.diff == 1:
if "left" in bc.name:
res = nm.stack(((4*(x_2 - 1)*cos(4) - 5*x_2*cos(5*x_2))*
(exp(-t) - 1),
-5*(exp(-t) - 1)*cos(5*x_2)),
axis=-2)
elif "bottom" in bc.name:
res = nm.stack(((5*cos(-5*x_1) - 8*cos(8*x_1 - 4))*(exp(-t) - 1),
-(5*x_1*cos(-5*x_1) - 4*(x_1 - 1)*cos(8*x_1 - 4))*
(exp(-t) - 1)),
axis=-2)
elif "right" in bc.name:
res = nm.stack(((4*(x_2 - 1)*cos(4) - 5*x_2*cos(5*x_2))*
(exp(-t) - 1),
-5*(exp(-t) - 1)*cos(5*x_2)),
axis=-2)
elif "top" in bc.name:
res = nm.stack((-5*(exp(-t) - 1)*cos(5*x_1),
(4*(x_1 - 1)*cos(4) - 5*x_1*cos(5*x_1))*
(exp(-t) - 1)),
axis=-2)
return res
@local_register_function
def source_fun(ts, coors, mode="qp", **kwargs):
if mode == "qp":
t = ts.dt * ts.step
x_1 = coors[..., 0]
x_2 = coors[..., 1]
sin = nm.sin
cos = nm.cos
exp = nm.exp
res = (
+ (5 * x_1 * cos(5 * x_1 * x_2)
- 4 * (x_1 - 1) * cos(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2)) *
(exp(-t) - 1) ** 2 * (sin(5 * x_1 * x_2)
- sin(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2))
+ (5 * x_2 * cos(5 * x_1 * x_2)
- 4 * (x_2 - 1) * cos(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2)) *
(exp(-t) - 1) ** 2 * (sin(5 * x_1 * x_2)
- sin(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2))
- diffcoef *
((25 * x_1 ** 2 * sin(5 * x_1 * x_2) - 16 * (x_1 - 1) ** 2 *
sin(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2)) * (exp(-t) - 1)
+ (25 * x_2 ** 2 * sin(5 * x_1 * x_2) - 16 * (x_2 - 1) ** 2 *
sin(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2)) * (exp(-t) - 1))
+ (sin(5 * x_1 * x_2) - sin(4 * x_1 * x_2 - 4 * x_1 - 4 * x_2))*
exp(-t)
)
return {"val": res[..., None, None]}
def adv_fun(p):
vu = velo.T * p[..., None]
return vu
def adv_fun_d(p):
v1 = velo.T * nm.ones(p.shape + (1,))
return v1
def burg_fun(p):
vu = .5*burg_velo * p[..., None] ** 2
return vu
def burg_fun_d(p):
v1 = burg_velo * p[..., None]
return v1
materials = {
'a' : ({'val': [velo], '.flux':adflux},),
'D' : ({'val': [diffcoef], '.Cw': cw},),
'g' : 'source_fun'
}
ics = {
'ic': ('Omega', {'p.0': 0}),
}
dgebcs = {
'u_left' : ('left', {'p.all': 'bc_funs', 'grad.p.all': 'bc_funs'}),
'u_right' : ('right', {'p.all': 'bc_funs', 'grad.p.all': 'bc_funs'}),
'u_bottom' : ('bottom', {'p.all': 'bc_funs', 'grad.p.all': 'bc_funs'}),
'u_top' : ('top', {'p.all': 'bc_funs', 'grad.p.all': 'bc_funs'}),
}
equations = {
'balance':
"dw_dot.i.Omega(v, p)" +
# non-linear hyperbolic terms
" - dw_ns_dot_grad_s.i.Omega(burg_fun, burg_fun_d, p[-1], v)" +
" + dw_dg_nonlinear_laxfrie_flux.i.Omega(a.flux, burg_fun, burg_fun_d, v, p[-1])" +
# diffusion
" + dw_laplace.i.Omega(D.val, v, p[-1])" +
diffusion_schemes_explicit[diffscheme] +
" - dw_dg_interior_penalty.i.Omega(D.val, D.Cw, v, p[-1])"
# source
+ " - dw_volume_lvf.i.Omega(g.val, v)"
" = 0"
}
solvers = {
"tss.tvd_runge_kutta_3": ('ts.tvd_runge_kutta_3',
{"t0": t0,
"t1": t1,
'limiters': {
"f": MomentLimiter2D} if limit else {}}),
"tss.euler": ('ts.euler',
{"t0" : t0,
"t1" : t1,
'limiters': {"f": MomentLimiter2D} if limit else {}}),
'nls': ('nls.newton', {}),
'ls' : ('ls.scipy_direct', {})
}
options = {
'ts' : 'tss.euler',
'nls' : 'nls.newton',
'ls' : 'ls.mumps',
'save_times' : 100,
'output_dir' : 'output/dg/' + example_name,
'output_format' : 'msh',
'file_format' : 'gmsh-dg',
'pre_process_hook': get_cfl_setup(CFL=cfl, dt=dt)
}
return locals()